Introduction
Design of Experimen refers to the process of planning the experiment so that the appropriate data will be collected which may be analyzed by a statistical method. Design and analysis of experiments is a branch of statistics that is crucial and is applied in various sectors and industries such as health, agrigiculture and engineering.
There are may designs of experiments; Completely Randomized Design (CRD) Randomized Bleck Design (RBD) Latin Sqaure Designs (LSD) Factorial Experiments
In this blog, we will discuss performing CRD in R
Completely Randomized Design
This design contains one factor and thus only one way classification the general staistical model is: Yij=µ+αi+εij,
Where:
µ is the overall(grand) mean common to all treatment. It is the mean yield in the absence of the treatment effect of the experiment.
σ²εij is the random error of the jth observation receiving the ith treatment and is assumed to be N~iid(0, σ²)
εij is the random error of the jth observation receiving the ith treatment and is assumed to be N~iid(0, σ²)
The hypothesis under test is:
Null- αi=0, for all i
Alternative- αi≠0, for any i
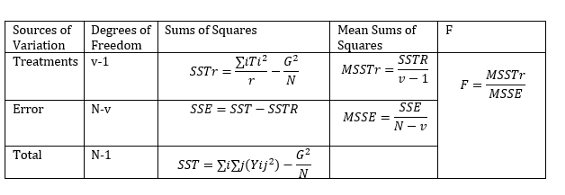
To perform manual ANOVA we follow the table below
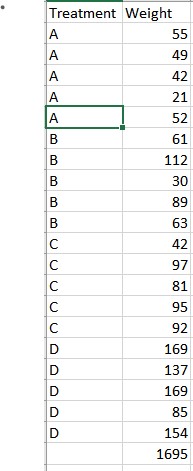
Find the dataset below; https://github.com/StatisticianLeboo/statistics-topics/tree/ANOVA
CRD
Example 1
The data contains 4 tropical food stuffs A, B, C, and D tried on 20 chicks. Analyze the data at 5% level of significance.
Solution
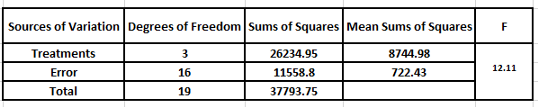 We compare the computed F value with the tabulated one from the F tables 5% (3,16)=3.24. Since the tabulated value is small you reject the null hypothesis and conclude that the treatments vary significantly and do not have similar effect on the chicks.
We compare the computed F value with the tabulated one from the F tables 5% (3,16)=3.24. Since the tabulated value is small you reject the null hypothesis and conclude that the treatments vary significantly and do not have similar effect on the chicks.
An additional test; LSD is used to find which of the foods is mostly significant.
In R, we perform the analysis as follows:
library(readxl)
chicks <- read_excel("chics.xlsx")
 Set the treatment column into factors
Set the treatment column into factors
chicks$Treatment<-as.factor(chicks$Treatment)
Performing ANOVA
Method 1
mod1<-aov(Weight~Treatment, data = chicks)
summary(mod1)
## Df Sum Sq Mean Sq F value Pr(>F)
## Treatment 3 26235 8745 12.11 0.000218 ***
## Residuals 16 11559 722
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## 5 observations deleted due to missingness
Method 2
chicksmodel<-lm(Weight~Treatment, data = chicks)
anova(chicksmodel)
## Analysis of Variance Table
##
## Response: Weight
## Df Sum Sq Mean Sq F value Pr(>F)
## Treatment 3 26235 8745.0 12.105 0.000218 ***
## Residuals 16 11559 722.4
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
LSD Test
library(agricolae)
LSD.test(chicksmodel,"Treatment", console = TRUE)
##
## Study: chicksmodel ~ "Treatment"
##
## LSD t Test for Weight
##
## Mean Square Error: 722.425
##
## Treatment, means and individual ( 95 %) CI
##
## Weight std r LCL UCL Min Max
## A 43.8 13.62718 5 18.31833 69.28167 21 55
## B 71.0 31.02418 5 45.51833 96.48167 30 112
## C 81.4 22.87575 5 55.91833 106.88167 42 97
## D 142.8 34.90272 5 117.31833 168.28167 85 169
##
## Alpha: 0.05 ; DF Error: 16
## Critical Value of t: 2.119905
##
## least Significant Difference: 36.03652
##
## Treatments with the same letter are not significantly different.
##
## Weight groups
## D 142.8 a
## C 81.4 b
## B 71.0 bc
## A 43.8 c
From the LSD, it is clear that treatment D is the most effective. There is no much difference between C and B, whereas A has the least treatment effect.
Example 2
The data contains the amount of electricity used in KWh in three towns. Test at 5% significance whether the amount of electricity used is the same in the 3 towns. The steps are as example 1
library(readxl)
electricity <- read_excel("electricity.xlsx")
#Set the treatment variable to factor
electricity$Town<-as.factor(electricity$Town)
model2<-aov(Electricity_Used~Town, data=electricity)
summary(model2)
## Df Sum Sq Mean Sq F value Pr(>F)
## Town 2 150.6 75.30 2.705 0.085 .
## Residuals 27 751.6 27.84
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1