Introduction to Factorial Experiments
This is the last section on my series of the various designs of experiments using R. These designs are what are commonly referred to as one factor at a time experiment. Make sure to read the previous post on CRD and RBD experiments.
What are Factorial Experiments?
Many experiments involve the study of the effects of two or more factors. In general, factorial designs are most efficient for this type of experiment. By a factorial design, we mean that in each complete trial or replicate of the experiment all possible combinations of the levels of the factors are investigated. For example, if there are a levels of factor A and b levels of factor B, each replicate contains all ab treatment combinations. When factors are arranged in a factorial design, they are often said to be crossed. The effect of a factor is defined to be the change in response produced by a change in the level of the factor. This is frequently called a main effect because it refers to the primary factors of interest in the experiment. (Montgomery, 2012)
Factorial experiments allow us to analyze the effects of factors simultaneously. The most common are the 2 factor factorials also known as the 2k experiments, where k can take the values (2,3,…).
Advantages of Factorial Experiments Over One Factor at a Time Experiments
- They are more efficient in that they provide more information at lower or similar cost as compared to one factor at a time experiments.
- Allows a researcher to study how the factors interact within themselves.
Disadvantages
- Some experiments may be cumbersome to compute mathematically especially if they involve numerous factors.
- Some information may be lost during the design of a factorial experiment. This is a phenomenon known as confounding.
I will not get into the manual derivations of how to solve factorial experiments but, I will however attach some reference where one can get more information about it.
Factorial Designs Using R
To perform these experiments, we will use the DoeBioresearch package. Additionally, I have partnered with two friends; who are both microbiologists and have allowed me to use their experimental data for this illustration.
Example 1
In this example, my friend Joseah Lang’at check out his LinkedIn profile was performing an experiment SENSITIVITY OF *Staphylococcus aureus AND Escherichia coli TO Zingiber officinale AND Allium sativum EXTRACTS*. In simpler terms, the title of the experiment was assessing how sensitive the two bacteria were to extracts of ginger and garlic.
The main objectives of the experiment were; To determine the sensitivity of Staphylococcus aureus and Escherichia coli to Zingiber officinale and Allium sativum.
The data contained the following; Concentration. This represents levels of distilled water. (1,2,3) representing (0%, 50%, 25%) distilled water respectively. Treatments. There Value. This is the zone of inhibition recorded in the experiment. The values are in centimeters.
Loading the data and required packages
library(doebioresearch)
library(readxl)
sensitivity_data <- read_excel("SensitivityData.xlsx",
sheet = "saureus")
Some Visualizations
library(ggplot2)
library(ggthemes)
ggplot(sensitivity_data) +
aes(x = Treatment, y = Value, fill = Treatment) +
geom_boxplot(shape = "circle") +
scale_fill_hue(direction = 1) +
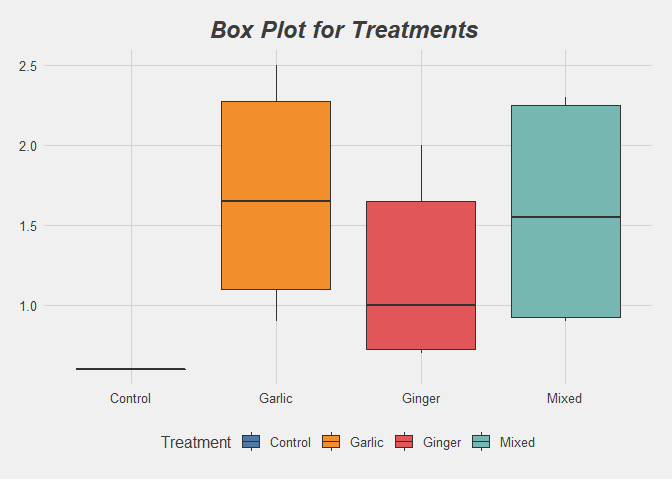
labs(title = "Box Plot for Treatments ") +
scale_fill_tableau()+
ggthemes::theme_fivethirtyeight() +
theme(plot.title = element_text(face = "bold.italic",hjust = 0.5))
ggplot(sensitivity_data) +
aes(x = Treatment, fill = Treatment, weight = Value) +
geom_bar() +
scale_fill_brewer(palette = "Set1", direction = 1) +
labs(y = "Yield",
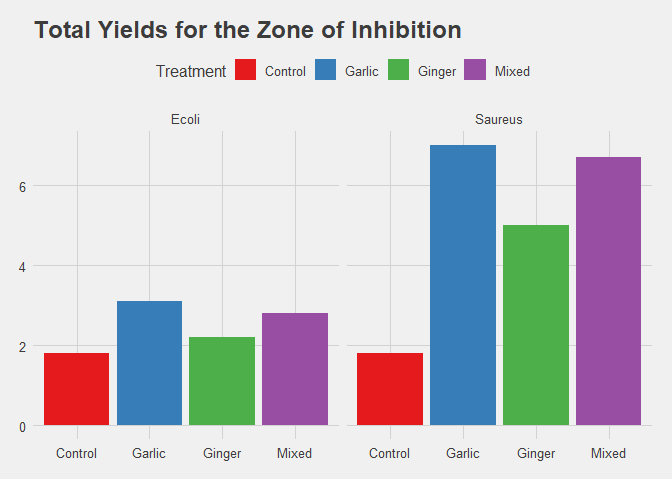
title = "Total Yields for the Zone of Inhibition") +
ggthemes::theme_fivethirtyeight() +
theme(legend.position = "top") +
facet_wrap(vars(Bacteria))
ggplot(sensitivity_data) +
aes(x = Bacteria, y = Value, fill = Bacteria) +
geom_boxplot(shape = "circle") +
scale_fill_hue(direction = 1) +
labs(title = "Boxplot for the Bacteria") +
theme_minimal() +
scale_fill_tableau()+
theme(plot.title = element_text(face = "bold.italic",hjust = 0.5))
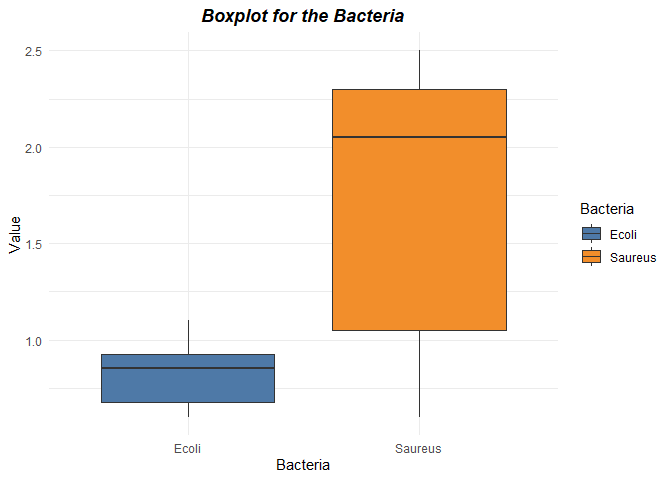
No presence of Outliers from the plots above.
Running a factorial experiment.
To analyze the significance of the factors in the experiment, a 2^2 factorial experiment was chosen. This experiment was carried out at 5% level of significance. The Least Significant Difference was used to check on the of the differences between statistically significant groups. The two factors are Bacteria and Treatments. Concentration was taken as the blocking/replication factor.
Hypotheses
- Treatments (Factor A) Null: Anti-microbial activity between the treatments are equal. Alternative: At least one of the treatments has a different Anti-microbial activity.
- Bacteria (Factor B) Null: S.aureus and E.coli have similar sensitivity effect on the extracts. Alternative: S.aureus and E.coli do not have similar sensitivity effect on the extracts.
- Blocks (Concetration of Distilled Water) Null: Blocks have similar effect Alternative: At least one is different
Decision Rule If p-value is less than alpha(0.05) we reject the null hypothesis.
frbd2fact(sensitivity_data[3], sensitivity_data$Concentration,
sensitivity_data$Treatment,sensitivity_data$Bacteria,1)
## $Value
## $Value[[1]]
## Analysis of Variance Table
##
## Response: dependent.var
## Df Sum Sq Mean Sq F value Pr(>F)
## replicationvector 2 0.1408 0.0704 3.0888 0.07741 .
## fact.A 3 4.3367 1.4456 63.4082 2.167e-08 ***
## fact.B 1 4.6817 4.6817 205.3577 9.299e-10 ***
## fact.A:fact.B 3 1.6950 0.5650 24.7833 7.315e-06 ***
## Residuals 14 0.3192 0.0228
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## $Value[[2]]
## [1] "R Square 0.971"
##
## $Value[[3]]
## [1] "SEm of A: 0.062 , SEd of A: 0.087 , SEm of B: 0.044 , SEd of B 0.062 , SEm of AB: 0.087 , SEd of AB: 0.123"
##
## $Value[[4]]
##
## Shapiro-Wilk normality test
##
## data: model$residuals
## W = 0.89877, p-value = 0.02027
##
## $Value[[5]]
## [1] "Normality assumption is violated"
##
## $Value[[6]]
## [1] "The means of one or more levels of first factor are not same, so go for multiple comparison test"
##
## $Value[[7]]
## $Value[[7]][[1]]
## MSerror Df Mean CV t.value LSD
## 0.02279762 14 1.266667 11.92017 2.144787 0.1869684
##
## $Value[[7]][[2]]
## dependent.var groups
## Garlic 1.683333 a
## Mixed 1.583333 a
## Ginger 1.200000 b
## Control 0.600000 c
##
## $Value[[8]]
## [1] "The means of one or more levels of second factor are not same, so go for multiple comparison test"
##
## $Value[[9]]
## $Value[[9]][[1]]
## MSerror Df Mean CV t.value LSD
## 0.02279762 14 1.266667 11.92017 2.144787 0.1322066
##
## $Value[[9]][[2]]
## dependent.var groups
## Saureus 1.708333 a
## Ecoli 0.825000 b
##
## $Value[[10]]
## [1] "The means of levels of interaction between two factors are not same, so go for multiple comparison test"
##
## $Value[[11]]
## $Value[[11]][[1]]
## MSerror Df Mean CV t.value LSD
## 0.02279762 14 1.266667 11.92017 2.144787 0.2644133
##
## $Value[[11]][[2]]
## dependent.var groups
## Garlic:Saureus 2.3333333 a
## Mixed:Saureus 2.2333333 a
## Ginger:Saureus 1.6666667 b
## Garlic:Ecoli 1.0333333 c
## Mixed:Ecoli 0.9333333 cd
## Ginger:Ecoli 0.7333333 de
## Control:Ecoli 0.6000000 e
## Control:Saureus 0.6000000 e
Result Interpretation
- ANOVA TABLE
Are the concentration (Block) significant?
The test returned a p-value of (0.07741) for the blocks. This is greater than 0.05. The decision is therefore fail to reject the null hypothesis and conclude that with 95% confidence, the concentration effects of distilled water are statistically insignificant to the experimental yield.
Are the Effects of the Extracts Significant?
A p-value of (2.167e-08) was returned by the test. This is less than 0.05. The null hypothesis is rejected and conclude that with 95% confidence, that the effects of the plant extracts differ significantly.
Are the Effects of the Bacteria Significant?
The p-value (9.299e-10) is less than 0.05. We therefore reject the null hypothesis and conclude that with 95% confidence, that the effects of the bacteria differ significantly. Main effects are significant
Are the Two-Factor Interaction Effects Significant?
The p-value (9.299e-10) is less than 0.05. We therefore reject the null hypothesis and conclude that with 95% confidence, that the interaction effects are statistically significant.
R Square 0.971, indicates that this model explains 97.1% variability. This is a good result.
- Multiple Comparison Tests Having rejected the null hypothesis in for main effects and interactions, the LSD test was used to test and select the effects with the most significance.
Treatments
Of the 4 treatments, no statistical difference exists between Garlic and Mixed. These two offer the highest anti-microbial activity. When compared across the treatments, Garlic and Mixed differed significantly from ginger and control.
Bacteria
S.aureus had the highest sensitivity as compared to E.coli with a mean of 1.708 as compared to 0.825 for E.coli.
Interaction Effects
The interaction Garlic:S.aureus and Mixed:S.aureus offered the highest yield and there was no statistical differences between these interactions.
In chronological order, the interaction effects were as follows;
- Garlic:Saureus and Mixed:Saureus
- Ginger:Saureus
- Garlic:Ecoli
- Mixed:Ecoli
- Ginger:Ecoli
- Control:Ecoli
7.Control:Saureus
To conclude this experiment, an interaction plot was used to visualize the presecence of interaction between the two factors.
Checking for interaction within the data
We use the interaction.plot function to check on interaction between the two factors; Bacteria and treatment
interaction.plot(x.factor=sensitivity_data$Bacteria,
trace.factor=sensitivity_data$Treatment,
response=sensitivity_data$Value,
type="b",
main="Interaction Between Bacteria and Plant Extracts",
xlab="Bacteria",
ylab = "Zone of Inhibition in cm",
col = c("red","blue", "green", "black"),
lwd=2,
trace.label = "Extract")
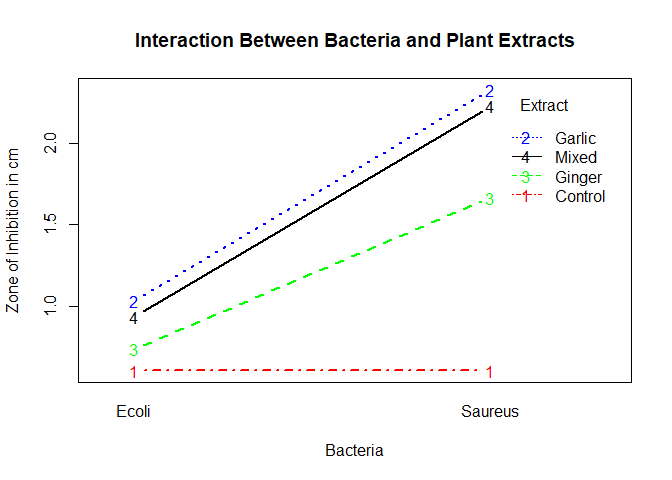 From the plot, Garlic has the highest antimicrobial activity followed by mixed then ginger and finally control. S.aureus has higher sensitivity as compared to E.coli. However, we can not conclude that there is presence of interaction between the two factors. Additionally, there is an increase in the sensitivity of the bacteria as you move from E.coli
to S.aureus
From the plot, Garlic has the highest antimicrobial activity followed by mixed then ginger and finally control. S.aureus has higher sensitivity as compared to E.coli. However, we can not conclude that there is presence of interaction between the two factors. Additionally, there is an increase in the sensitivity of the bacteria as you move from E.coli
to S.aureus
Example 2
In this example, my friend Gideon check out his [LinkedIn page]9(https://www.linkedin.com/in/steve-gideons-b390761a1/) was performing an experiment title Prevalence of Bovine Mastitis and their Anti-microbial Susceptibility Profile in London Ward, Nakuru County The objectives of the research were to determine the prevelence of mastitis, identify the most prevalent bacteria causing mastitis and determine their antimicrobial susceptibility profile.
Bovine Mastitis is an inflammatory response of the udder tissue in mammary gland caused due to physical trauma or microorganism infections. It is considered the most common disease leading to economic loss in dairy industries due to reduced yield and poor quality of milk. (Henriques and Gomes, 2016)
In summary, the research sought to investigate the most dominant bacteria that causes bovine mastitis and which type of mastitis diagnosis is the most prevalent. Finally, the research looked into some common antibiotics to understand which one was effective against what type of bovine mastitis. The study was conducted in London ward and a sample of 15 cows were randomly selected and milk samples drawn from them. Laboratory tests were performed and respective zone of inhibition recorded.
Loading the Data and some required packages
library(readxl)
library(dplyr)
library(tidyr)
library(knitr)
library(ggplot2)
library(ggthemes)
Mastistis <- read_excel("MastitisData.xlsx",
sheet = "Sheet1", col_types = c("text",
"text", "text", "text", "text", "text",
"numeric"))
The data contained the following variables;
- Specimen_No
- Breed - Type of cow; Friesian, Jersey or Ayshire
- Quarters - Cow’s tits. Front Left (FL), Front Right, Back Left, Back Right
- Isolates - Bacteria identified
- Diagnosis - Type of mastitis
- Sensitivity_test - Antibiotic administered
- Yield - Zone of inhibition recorded in millimeters.
Some Exploratory Data Analysis
Zone of Inhibition by Breed
Breed_Summary<-Mastistis %>%
group_by(Breed) %>%
drop_na() %>%
summarise(Count=n(),
Average_Zone= mean(Yield),
SD=sd(Yield),
SE=SD/Count,
LCL=round(Average_Zone-(1.96*SE),2),
UCL=round(Average_Zone+(1.96*SE),2),
Zone_Mean_95Pct_CI=paste("(", LCL ," , ", UCL, ")" ))
kable(Breed_Summary %>%
select(-c(LCL,UCL, SE)))
| Breed | Count | Average_Zone | SD | Zone_Mean_95Pct_CI |
|---|---|---|---|---|
| Ayshire | 80 | 19.65000 | 5.388924 | ( 19.52 , 19.78 ) |
| Fresian | 168 | 19.10119 | 5.439531 | ( 19.04 , 19.16 ) |
| Jersey | 56 | 19.73214 | 4.669012 | ( 19.57 , 19.9 ) |
Aggregation by Isolates
Isolate<-Mastistis %>%
group_by(Isolates) %>%
summarise(Count=n()) %>%
mutate(Pct=(round(Count/sum(Count),2)*100),
Isolate_int=c("Ent","E.coli", "Klebsiella", "No_Isolate", "Pseudomonas",
"Staphylococcus"),
Rate=paste0(Pct, "%"),
cumulative_ratio=cumsum(Pct),
ration_min=c(0, head(cumulative_ratio,n=-1)),
labelPosition=(cumulative_ratio+ration_min)/2,
label=paste0(Isolate_int, ":", Rate))
ggplot(Isolate,aes(ymax=cumulative_ratio, ymin=ration_min, xmax=4, xmin=3,
fill=Isolate_int))+
geom_rect()+
scale_fill_manual(values = c(Ent = "red",
E.coli = "orange",
Klebsiella= "green",
No_Isolate = "blue",
Pseudomona = "yellow",
Staphylococcus= "purple"))+
geom_label(x=2.6,aes(y=labelPosition, label=label),size=6)+
coord_polar(theta="y")+
xlim(c(-1,4))+
labs(title="ISOLATES",
subtitle = ("Enterecoccus, E.coli, Klebsiella, No_Isolate, Pseudomonas,
Staphylococcus"))+
theme(plot.title = element_text(face = "bold.italic",hjust = 0.5))+
theme_void()+
theme(legend.position = "none")
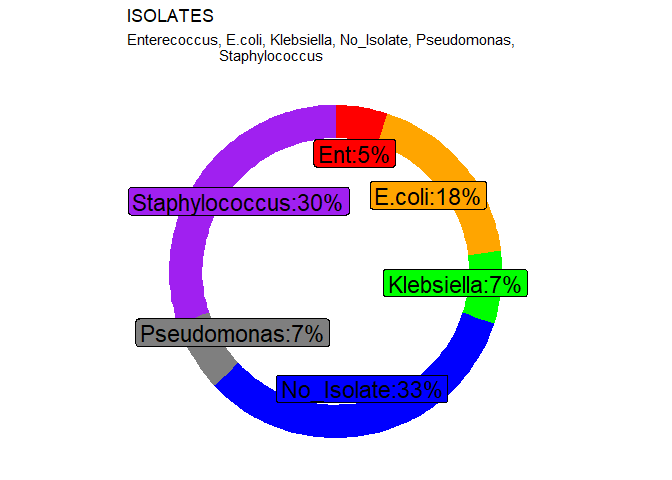 33% of the samples showed no isolate. This is a warying statistic since it implies that the prevalence rate of Bovine mastitis is at 67% which is relatively high.
33% of the samples showed no isolate. This is a warying statistic since it implies that the prevalence rate of Bovine mastitis is at 67% which is relatively high.
Dropping values with No isolates
no_isolate<-Mastistis %>%
filter(Isolates=="No isolate")
Mastistis_clean<-Mastistis[!Mastistis$Isolates=="No isolate",]
Mastistis_clean <-Mastistis_clean %>%
mutate(Breed=as.factor(Breed),
Quarters=as.factor(Quarters),
Isolates=as.factor(Isolates),
Diagnosis=as.factor(Diagnosis),
Yield=as.numeric(Yield))
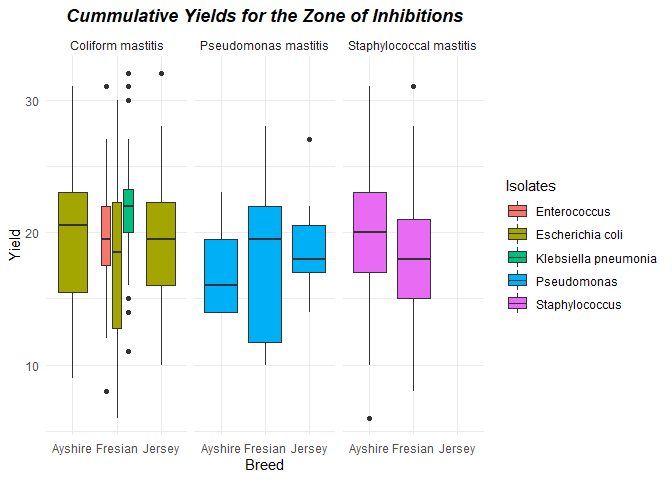
Cummulative Yields for the Zone of Inhibitions
ggplot(Mastistis_clean) +
aes(x = Breed, y = Yield, fill = Isolates) +
geom_boxplot(shape = "circle") +
scale_fill_hue(direction = 1) +
labs(title = "Cummulative Yields for the Zone of Inhibitions") +
theme_minimal() +
theme(plot.title = element_text(face = "bold.italic",hjust = 0.5)) +
facet_wrap(vars(Diagnosis))
Analysis of Diagnosis and the Anti-microbial Agents
To test for the significance in the mean differences, 3k factorial experiment was chosen.The Least Significant Difference was used to check on the of the differences between statistically significant groups. The three factors are Diagnosis, Sensitivity and Breed. Quarters was taken as the blocking/replication factor.
Hypotheses
-
Diagnosis (Factor A) Null: The diagnosis are similar in effect. Alternative: At least one of the diagnosis has a different effect.
-
Sensitivity (Factor B) These are the drugs subjected to the pathogens associated with mastitis. Null: The Sensitivity have similar effects. Alternative: At least one of the Sensitivity is different.
-
Breed (Factor C) Null: The breeds have similar effects. Alternative: At least one of the breeds is different.
-
Blocks Null: Blocks have similar effect. Alternative: At least one is different.
-
Interaction effects Null: Two and three factor interactions have similar effect. Alternative: At least one the interaction sets is different.
Decision rule p-value criteria. If p-value is less than alpha (0.05), the null hypothesis is rejected.
library(doebioresearch)
frbd3fact(Mastistis_clean[7], Mastistis_clean$Quarters, Mastistis_clean$Diagnosis,
Mastistis_clean$Sensitivity_test, Mastistis_clean$Breed, 1)
## $Yield
## $Yield[[1]]
## Analysis of Variance Table
##
## Response: dependent.var
## Df Sum Sq Mean Sq F value Pr(>F)
## replicationvector 9 179.2 19.909 0.9633 0.471279
## fact.A 2 220.9 110.441 5.3434 0.005388 **
## fact.B 7 1405.0 200.720 9.7113 1.366e-10 ***
## fact.C 2 1.5 0.728 0.0352 0.965412
## fact.A:fact.B 14 589.4 42.098 2.0368 0.016189 *
## fact.A:fact.C 3 41.6 13.854 0.6703 0.571066
## fact.B:fact.C 14 729.8 52.129 2.5221 0.002284 **
## fact.A:fact.B:fact.C 21 518.4 24.688 1.1945 0.256982
## Residuals 231 4774.5 20.669
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## $Yield[[2]]
## [1] "R Square 0.436"
##
## $Yield[[3]]
## [1] "SEm of A: 0.293 , SEd of A: 0.415 , SEm of B: 0.479 , SEd of B 0.678 , SEm of C: 0.293 , SEd of C: 0.415 , SEm of AB: 0.83 , SEd of AB: 1.174 , SEm of AC: 0.508 , SEd of AC: 0.719 , SEm of BC: 0.83 , SEd of BC: 1.174 , SEm of ABC: 1.438 , SEd of ABC: 2.033"
##
## $Yield[[4]]
##
## Shapiro-Wilk normality test
##
## data: model$residuals
## W = 0.98536, p-value = 0.003476
##
##
## $Yield[[5]]
## [1] "Normality assumption is violated"
##
## $Yield[[6]]
## [1] "The means of one or more levels of factor A are not same, so go for multiple comparison test"
##
## $Yield[[7]]
## $Yield[[7]][[1]]
## MSerror Df Mean CV
## 20.66862 231 19.36184 23.48059
##
## $Yield[[7]][[2]]
## dependent.var groups
## Coliform mastitis 20.05882 a
## Staphylococcal mastitis 18.92647 b
## Pseudomonas mastitis 18.25000 b
##
##
## $Yield[[8]]
## [1] "The means of one or more levels of factor B are not same, so go for multiple comparison test"
##
## $Yield[[9]]
## $Yield[[9]][[1]]
## MSerror Df Mean CV t.value LSD
## 20.66862 231 19.36184 23.48059 1.970287 2.054983
##
## $Yield[[9]][[2]]
## dependent.var groups
## Gentamycin 23.31579 a
## Neomycin 20.68421 b
## Cefalexin 20.57895 b
## Streptomycin 19.60526 bc
## Tetracycline 18.73684 bc
## Kanamycin 18.71053 bc
## Penicillin 17.71053 c
## Cotrimoxazole 15.55263 d
##
##
## $Yield[[10]]
## [1] "All the factor C level means are same so dont go for any multiple comparison test"
##
## $Yield[[11]]
## $Yield[[11]][[1]]
## MSerror Df Mean CV
## 20.66862 231 19.36184 23.48059
##
## $Yield[[11]][[2]]
## dependent.var groups
## Jersey 19.73214 a
## Ayshire 19.65000 a
## Fresian 19.10119 a
##
## $Yield[[12]]
## [1] "The means of levels of interaction between A and B factors are not same, so go for multiple comparison test"
##
## $Yield[[13]]
## $Yield[[13]][[1]]
## MSerror Df Mean CV
## 20.66862 231 19.36184 23.48059
##
## $Yield[[13]][[2]]
## dependent.var groups
## Coliform mastitis:Gentamycin 24.05882 a
## Staphylococcal mastitis:Gentamycin 23.70588 ab
## Coliform mastitis:Tetracycline 21.64706 abc
## Coliform mastitis:Cefalexin 21.58824 abc
## Pseudomonas mastitis:Neomycin 21.50000 abcd
## Staphylococcal mastitis:Neomycin 20.76471 bcd
## Pseudomonas mastitis:Kanamycin 20.75000 bcde
## Coliform mastitis:Streptomycin 20.47059 cde
## Staphylococcal mastitis:Kanamycin 20.47059 cde
## Coliform mastitis:Neomycin 20.41176 cde
## Staphylococcal mastitis:Cefalexin 19.88235 cde
## Pseudomonas mastitis:Streptomycin 19.75000 cde
## Pseudomonas mastitis:Cefalexin 19.25000 cdef
## Coliform mastitis:Penicillin 18.70588 cdef
## Staphylococcal mastitis:Streptomycin 18.70588 cdef
## Pseudomonas mastitis:Gentamycin 18.50000 cdefg
## Pseudomonas mastitis:Tetracycline 17.75000 cdefg
## Coliform mastitis:Cotrimoxazole 17.11765 defg
## Staphylococcal mastitis:Penicillin 17.05882 defg
## Coliform mastitis:Kanamycin 16.47059 efg
## Pseudomonas mastitis:Penicillin 16.25000 efg
## Staphylococcal mastitis:Tetracycline 16.05882 efg
## Staphylococcal mastitis:Cotrimoxazole 14.76471 fg
## Pseudomonas mastitis:Cotrimoxazole 12.25000 g
##
##
## $Yield[[14]]
## [1] "The means of levels of interaction between B and C factors are not same, so go for multiple comparison test"
##
## $Yield[[15]]
## $Yield[[15]][[1]]
## MSerror Df Mean CV
## 20.66862 231 19.36184 23.48059
##
## $Yield[[15]][[2]]
## dependent.var groups
## Tetracycline:Jersey 26.14286 a
## Gentamycin:Ayshire 24.80000 ab
## Gentamycin:Fresian 23.23810 abc
## Cefalexin:Ayshire 21.50000 bcd
## Gentamycin:Jersey 21.42857 bcd
## Neomycin:Jersey 21.28571 bcd
## Streptomycin:Fresian 21.04762 cd
## Neomycin:Fresian 20.90476 cd
## Cefalexin:Fresian 20.85714 cd
## Kanamycin:Ayshire 20.30000 cd
## Neomycin:Ayshire 19.80000 cde
## Penicillin:Jersey 19.57143 cdef
## Kanamycin:Fresian 19.00000 def
## Tetracycline:Ayshire 18.90000 def
## Cefalexin:Jersey 18.42857 defg
## Streptomycin:Ayshire 18.30000 defg
## Cotrimoxazole:Jersey 18.28571 defg
## Penicillin:Ayshire 18.20000 defg
## Streptomycin:Jersey 17.14286 defg
## Penicillin:Fresian 16.85714 efg
## Tetracycline:Fresian 16.19048 fg
## Kanamycin:Jersey 15.57143 fg
## Cotrimoxazole:Ayshire 15.40000 fg
## Cotrimoxazole:Fresian 14.71429 g
##
##
## $Yield[[16]]
## [1] "The means of levels of interaction between A and C factors are same so dont go for any multiple comparison test"
##
## $Yield[[17]]
## $Yield[[17]][[1]]
## MSerror Df Mean CV
## 20.66862 231 19.36184 23.48059
##
## $Yield[[17]][[2]]
## dependent.var groups
## Coliform mastitis:Fresian 20.30556 a
## Staphylococcal mastitis:Ayshire 20.01786 a
## Coliform mastitis:Jersey 19.83333 a
## Coliform mastitis:Ayshire 19.62500 ab
## Pseudomonas mastitis:Jersey 19.12500 ab
## Pseudomonas mastitis:Fresian 18.37500 ab
## Staphylococcal mastitis:Fresian 18.16250 b
## Pseudomonas mastitis:Ayshire 17.12500 b
##
##
## $Yield[[18]]
## [1] "The means of levels of interaction between all the three factors ABC are same so dont go for any multiple comparison test"
##
## $Yield[[19]]
## $Yield[[19]][[1]]
## MSerror Df Mean CV
## 20.66862 231 19.36184 23.48059
##
## $Yield[[19]][[2]]
## dependent.var groups
## Pseudomonas mastitis:Neomycin:Fresian 27.00000 a
## Pseudomonas mastitis:Tetracycline:Jersey 27.00000 ab
## Coliform mastitis:Tetracycline:Jersey 26.00000 ab
## Staphylococcal mastitis:Gentamycin:Ayshire 25.57143 ab
## Coliform mastitis:Gentamycin:Fresian 25.44444 ab
## Coliform mastitis:Cefalexin:Ayshire 25.00000 ab
## Coliform mastitis:Gentamycin:Ayshire 24.00000 abc
## Pseudomonas mastitis:Kanamycin:Ayshire 23.00000 abcd
## Coliform mastitis:Cefalexin:Fresian 22.77778 abcd
## Coliform mastitis:Streptomycin:Fresian 22.66667 abcd
## Coliform mastitis:Penicillin:Ayshire 22.50000 abcd
## Staphylococcal mastitis:Gentamycin:Fresian 22.40000 abcd
## Coliform mastitis:Gentamycin:Jersey 22.00000 abcd
## Pseudomonas mastitis:Penicillin:Jersey 22.00000 abcde
## Coliform mastitis:Neomycin:Jersey 21.83333 abcde
## Pseudomonas mastitis:Cefalexin:Fresian 21.50000 abcde
## Pseudomonas mastitis:Streptomycin:Fresian 21.50000 abcde
## Staphylococcal mastitis:Cefalexin:Ayshire 21.14286 abcde
## Pseudomonas mastitis:Gentamycin:Ayshire 21.00000 abcdef
## Staphylococcal mastitis:Neomycin:Fresian 21.00000 abcdef
## Staphylococcal mastitis:Tetracycline:Ayshire 21.00000 abcdef
## Staphylococcal mastitis:Kanamycin:Ayshire 20.57143 abcdef
## Coliform mastitis:Neomycin:Ayshire 20.50000 abcdef
## Coliform mastitis:Streptomycin:Ayshire 20.50000 abcdef
## Coliform mastitis:Tetracycline:Fresian 20.44444 abcdef
## Staphylococcal mastitis:Neomycin:Ayshire 20.42857 abcdef
## Staphylococcal mastitis:Kanamycin:Fresian 20.40000 abcdef
## Pseudomonas mastitis:Kanamycin:Fresian 20.00000 abcdef
## Pseudomonas mastitis:Kanamycin:Jersey 20.00000 abcdef
## Staphylococcal mastitis:Streptomycin:Fresian 19.50000 bcdef
## Coliform mastitis:Neomycin:Fresian 19.44444 bcdef
## Coliform mastitis:Penicillin:Jersey 19.16667 bcdef
## Coliform mastitis:Cotrimoxazole:Jersey 19.00000 bcdef
## Pseudomonas mastitis:Streptomycin:Ayshire 19.00000 bcdef
## Staphylococcal mastitis:Cefalexin:Fresian 19.00000 bcdef
## Coliform mastitis:Cefalexin:Jersey 18.66667 bcdef
## Coliform mastitis:Kanamycin:Ayshire 18.00000 bcdef
## Pseudomonas mastitis:Gentamycin:Jersey 18.00000 bcdef
## Pseudomonas mastitis:Neomycin:Jersey 18.00000 bcdef
## Staphylococcal mastitis:Streptomycin:Ayshire 17.57143 bcdef
## Coliform mastitis:Penicillin:Fresian 17.55556 cdef
## Pseudomonas mastitis:Gentamycin:Fresian 17.50000 cdef
## Staphylococcal mastitis:Penicillin:Ayshire 17.42857 cdef
## Coliform mastitis:Kanamycin:Fresian 17.22222 cdef
## Coliform mastitis:Streptomycin:Jersey 17.16667 cdef
## Pseudomonas mastitis:Cefalexin:Ayshire 17.00000 cdef
## Pseudomonas mastitis:Cefalexin:Jersey 17.00000 cdef
## Pseudomonas mastitis:Streptomycin:Jersey 17.00000 cdef
## Coliform mastitis:Cotrimoxazole:Fresian 16.88889 def
## Staphylococcal mastitis:Penicillin:Fresian 16.80000 def
## Staphylococcal mastitis:Cotrimoxazole:Ayshire 16.42857 def
## Pseudomonas mastitis:Penicillin:Ayshire 15.00000 def
## Pseudomonas mastitis:Tetracycline:Fresian 15.00000 def
## Coliform mastitis:Kanamycin:Jersey 14.83333 def
## Coliform mastitis:Tetracycline:Ayshire 14.00000 def
## Pseudomonas mastitis:Cotrimoxazole:Ayshire 14.00000 def
## Pseudomonas mastitis:Cotrimoxazole:Jersey 14.00000 def
## Pseudomonas mastitis:Neomycin:Ayshire 14.00000 def
## Pseudomonas mastitis:Penicillin:Fresian 14.00000 def
## Pseudomonas mastitis:Tetracycline:Ayshire 14.00000 def
## Staphylococcal mastitis:Cotrimoxazole:Fresian 13.60000 ef
## Staphylococcal mastitis:Tetracycline:Fresian 12.60000 f
## Coliform mastitis:Cotrimoxazole:Ayshire 12.50000 f
## Pseudomonas mastitis:Cotrimoxazole:Fresian 10.50000 f
Interpretation of the ANOVA Results
Main Effects
-
Blocks The test reported a p-value of (0.471279) for the Quarters. This is greater than alpha (0.05). Decision Fail to reject the null hypothesis. Conclusion With 95% confidence, there exists sufficient statistical evidence to support the claim that the effects of the quarters are similar.
-
Factor A - Diagnosis The test reported a p-value of (0.005388) for the diagnosis. This is less than alpha (0.05). Decision Reject the null hypothesis. Conclusion With 95% confidence, there exists sufficient statistical evidence to deny the claim that the effects of the diagnosis are similar. The post hoc analyses showed that Coliform mastists had the highest effect with a mean prevalence of (20.05882). No statistical difference was found at 5% significance level between Staphylococcal mastitis and Pseudomonas mastitis.
-
Factor B - Sensitivity The test reported a p-value of (1.366e-10) for the Sensitivity. This is less than alpha (0.05). Decision Reject the null hypothesis. Conclusion With 95% confidence, there exists sufficient statistical evidence to deny the claim that the effects of the sensitivity are similar. The post hoc analyses showed that Gentamycin had the highest effect with a mean anti-microbial susceptibility of (23.31579). No statistical difference was found at 5% significance level between Neomycin and Cefalexin. Streptomycin, Tetracycline and Kanamycin had no statistical difference between them. Cotrimoxazole had the lowest anti-microbial susceptibility profile with mean (15.55263)
-
Factor C - Breed The test reported a p-value of (0.965412) for the breed. This is greater than alpha (0.05). Decision Fail to reject the null hypothesis. Conclusion With 95% confidence, there exists sufficient statistical evidence to support the claim that the effects of the breed are similar. This implies that the dignosis was not dependent on the breed.
Interaction Effect
-
fact.A:fact.B The two factor interaction for the diagnosis and sensitivity reported a p-value of (0.016189). This is less than alpha (0.05). Decision Reject the null hypothesis. Conclusion With 95% statistical confidence, there exists sufficient evidence to reject the claim that the interaction between diagnosis and sensitivity has similar effect. Post hoc LSD test reported Coliform mastitis:Gentamycin having the highest effect with mean (24.05882) implying that Coliform is highly susceptible to Gentamycin. No statistical difference was obtained between Coliform mastitis:Tetracycline and Coliform mastitis:Cefalexin. Pseudomonas mastitis:Cotrimoxazole offered the lowest effect with mean (12.25000).
-
fact.A:fact.C The two factor interaction for the diagnosis and breed reported a p-value of (0.571066). This is greater than alpha (0.05). Decision Fail to reject the null hypothesis. Conclusion With 95% statistical confidence, there exist sufficient statistical evidence to support the claim that the two factor interaction of diagnosis and breed have similar effects across all sets of combinations.
-
fact.B:fact.C The two factor interaction for the sensitivity and breed reported a p-value of (0.002284). This is less than alpha (0.05). Decision Reject the null hypothesis. Conclusion With 95% statistical confidence, there exists sufficient statistical evidence to reject the claim that the two factor interaction of sensitivity and breed have similar effects across all sets of combinations. Post hoc analyses showed that Tetracycline:Jersey exhibited the highest effect with mean (26.14286) while Cotrimoxazole:Fresian exhibited the lowest effect with mean (14.71429)
-
fact.A:fact.B:fact.C The three factor interaction reported a p-value of (0.256982). This greater than alpha (0.05). Decision Fail to reject the null hypothesis. Conclusion With 95% statistical confidence, there exists sufficient statistical evidence to support the claim that the three factor interaction of diagnosis, sensitivity and breed have similar effects across all sets of combinations.
Interaction plot
An interaction plot was generated to describe interaction between the two main factors; diagnosis and sensitivity.
interaction.plot(x.factor= Mastistis_clean$Diagnosis,
trace.factor=Mastistis_clean$Sensitivity_test,
response= Mastistis_clean$Yield,
type="b",
main="Interaction Between Diagnosis and Sensitivity Test",
xlab="Diagnosis",
ylab = "Zone of Inhibition in mm",
col = c("red","blue", "green", "violet", "gold", "brown", "black", "orange"),
lwd=3,
trace.label = "Sensitivity Test")
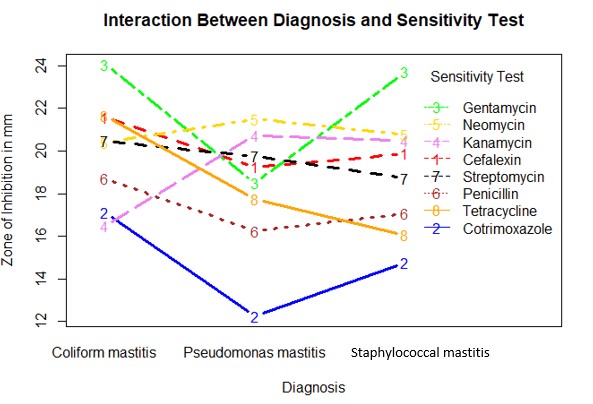
Interpretation
Gentamycin had the highest effects of all the sensitivity tests. Cotrimoxazole had the lowest effect. This could be attributed to high resistance of the pathogens to this anti-microbial agent.
There was an interaction of anti-microbial agents at different zones of inhibition. Cotrimoxazole and Kanamycin interacted at zone 17. Penicillin interacted with Kanamycin at zone 18 and Tetracycline at zone 17. Tetracyline interacted with Penicillin at zone 18, Kanamycin at zone 19, Streptomycin at zone 20. Neomycin at zone 21 and Cefalexin at zone 21. Gentamycin interacted with Neomycin at 21, Cefalexin at 21, Kanamycin at 20.
Moving across the diagnosis the study found the following; Gentamycin had highest effect on Coliform mastitis and Staphylococcal mastitis. There was a drop in the trend in Pseudomonas Mastitis indicating some of resistance. There was a general negative trend moving from Coliform mastitis to Pseudomonas Mastitis and a positive trend moving towards Staphylococcal mastitis for the sensitivity tests; Gentamycin, Cotrimoxazole, Penicillin and Cefalexin Tetracycline and Streptomycin exhibited a negative trend moving across Coliform mastitis and Staphylococcal mastitis. Kanamycin and Neomycin had a positive trend moving from Coliform mastitis to Pseudomonas mastitis. The trend was however relatively stable moving from Pseudomonas Mastitis to Staphylococcal mastitis
Conclusion
The factorial experiments are useful in that they allow us to analyze the effects of different factors simultaneously. The doebioresearch package contains numerous functions on how to analyze design of experiments data in biological research. We can analyze more than one dependent variable against various factors. Interaction plots visualize the interaction between to main factors. However, some may not clearly visualize the interaction. This does not mean that there are no interactions in the data.
References
Montgomery, D. C. (2012). Design and analysis of experiments. New York: John Wiley. Gomes F, Henriques M. Control of bovine mastitis: old and recent therapeutic approaches. Curr Microbiol. 2016;72:377–82. doi: 10.1007/s00284-015-0958-8. PubMed https://pubmed.ncbi.nlm.nih.gov/26687332/ Agricultural Statistics. Factorial RBD Analysis in 1 line along with LSD test in R [YouTube Video] https://youtu.be/8D2AFA3Dt8U Penn State University online lecture notes. https://online.stat.psu.edu/stat503/lesson/6